On July 31, the team of Guoqing Wang and Fengfeng Zhou of Jilin University published a paper titled “Application of Bayesian phylogenetic inference modelling for evolutionary genetic analysis and dynamic changes in 2019-nCoV” on the world-leading journal “Briefings in Bioinformatics”(Q1, IF=9.10),which analyzed the genome modification spectrum and molecular evolution characteristics of the novel coronavirus(2019-nCov) by combination of computational biology and molecular biology. The doctoral student Tong Shao and Master’s Degree student Wenfang Wang in the School of Basic Medical Sciences contributed equally to the article as co-first authors, while doctoral student Meiyu Duan in School of Computer Science assisted in the parallel analysis of massive viral data. Undergraduate student Baoyue Liu (class of 2016) in School of Pharmacy participated in this research through the College Student Innovation Practice Program. Professor Guoqing Wand from School of Basic Medical Sciences and Professor Fengfeng Zhou from School of Computer Science are the corresponding authors of this article. Jilin University School of Basic Medicine is the leading organization for this paper.
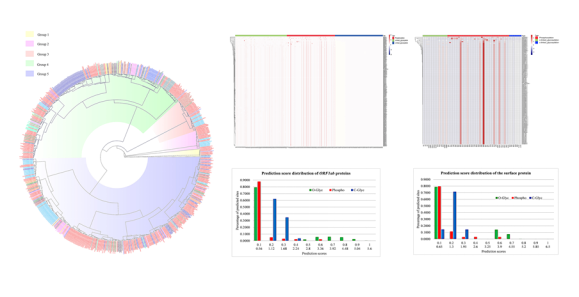
There have been more than 10 million cases of 2019-nCoV infection worldwide, and its rapid spread has caused catastrophic losses globally. In this study, the researchers used Bayesian methods to construct the evolutionary tree of 746 strains of 2019-nCoV, and analyzed the evolutionary relationship between virus strains from different regions. The strains from China and the United States belong to different branches, and their potential modification sites are different, suggesting that the strains in different regions may cause differences in infectivity and pathogenicity due to different modifications. Compared with SARS virus, 2019-nCoVORF6 has the lowest similarity (69%), and E protein has the highest similarity (95%). Compared with coronaviruses from bats, E protein and ORF6 (100%) have the highest similarity, and S protein has the lowest similarity (97%).
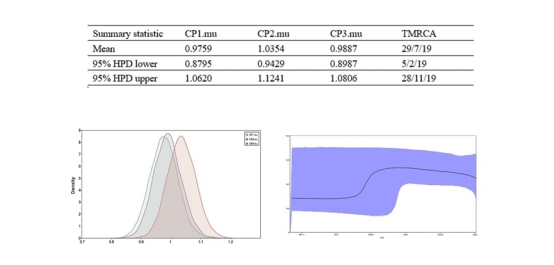
The evolutionary molecular clock showed that the common ancestor of 2019-nCoV appeared earlier than the first reported case. At the same time, the Bayesian skyline applied to estimate the active time of the effective population showed that its active time was also earlier than the first case, suggesting that 2019-nCoV may have existed for a long time. This working system analyzed the transmission mode and evolution direction of 2019-nCoV, and has important scientific significance for disease prevention and early warning.
This research was supported by the Ministry of Science and Technology "The Belt and Road Initiative" Infectious Disease Prevention and Control Science and Technology Major Project, the National Natural Science Foundation of China and the International Cooperation Project of Jilin Province Science and Technology Department.